Our research group is focused on understanding surveillance processes of the immune system, and the sabotage mechanisms used by pathogens to evade detection and host-mediated elimination.
We use the techniques of protein biochemistry, x-ray crystallography, cryo-EM, and computational biology to determine the three-dimensional structures of proteins and investigate their macromolecular interactions.
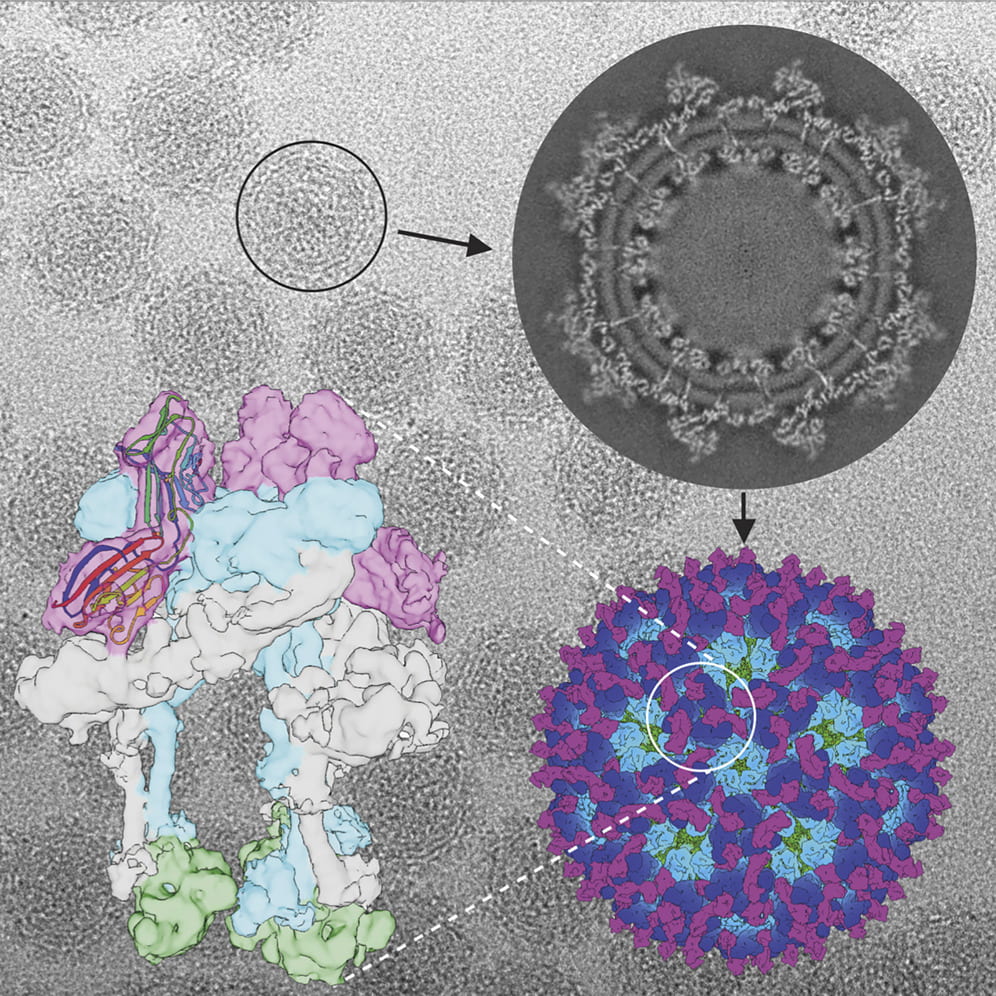
Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor
Basore K, Kim AS, Nelson CA, Zhang R, Smith BK, Uranga C, Vang L, Cheng M, Gross ML, Smith J, Diamond MS, Fremont DH. “Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor.” Cell. 2019 May 6. pii: S0092-8674(19)30392-7. doi: 10.1016/j.cell.2019.04.006.
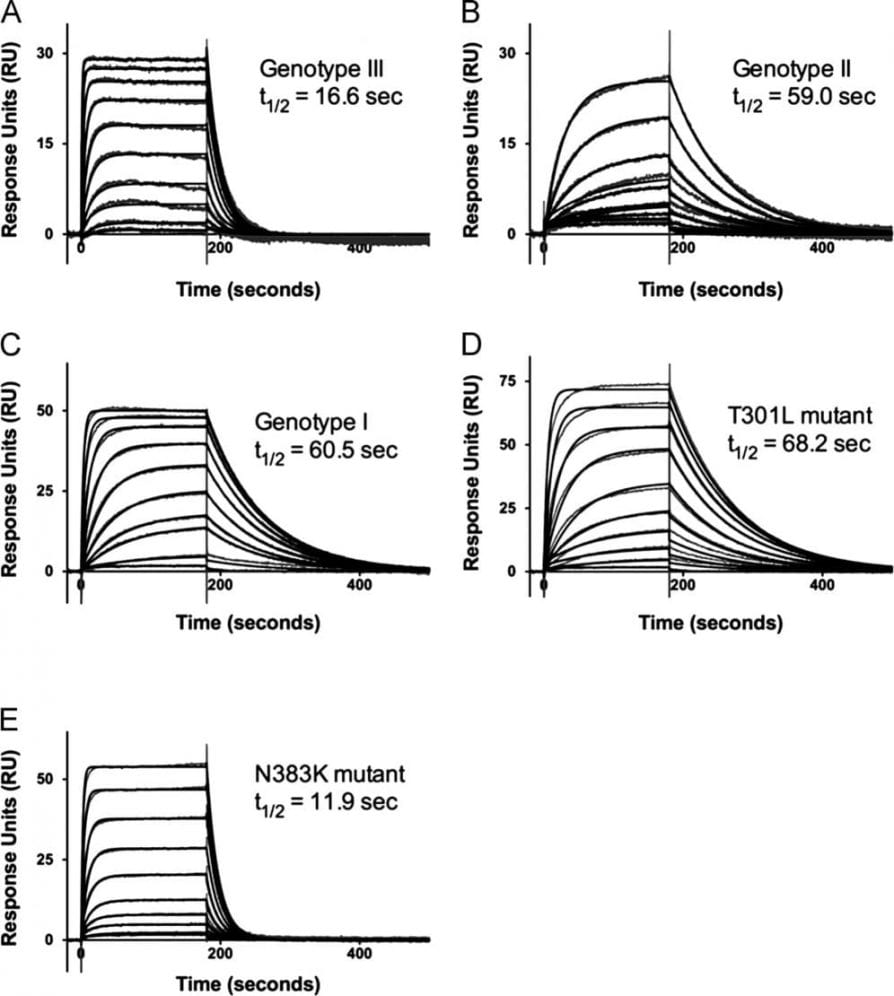
Kinetic analysis (using SPR) of 8A1 interaction with DENV-3 EDIII-MBP genotypes and mutants
Zhou Y, Austin SK, Fremont DH, Yount BL, Huynh JP, de Silva AM, Baric RS, Messer WB. “The mechanism of differential neutralization of dengue serotype 3 strains by monoclonal antibody 8A1.” Virology. 2013 Apr 25;439(1):57-64. doi: 10.1016/j.virol.2013.01.022.
Current interests of the lab include proteins associated with classical MHC antigen processing and presentation pathways, non-classical MHC family members, and natural killer cell activating and inhibitory receptors.
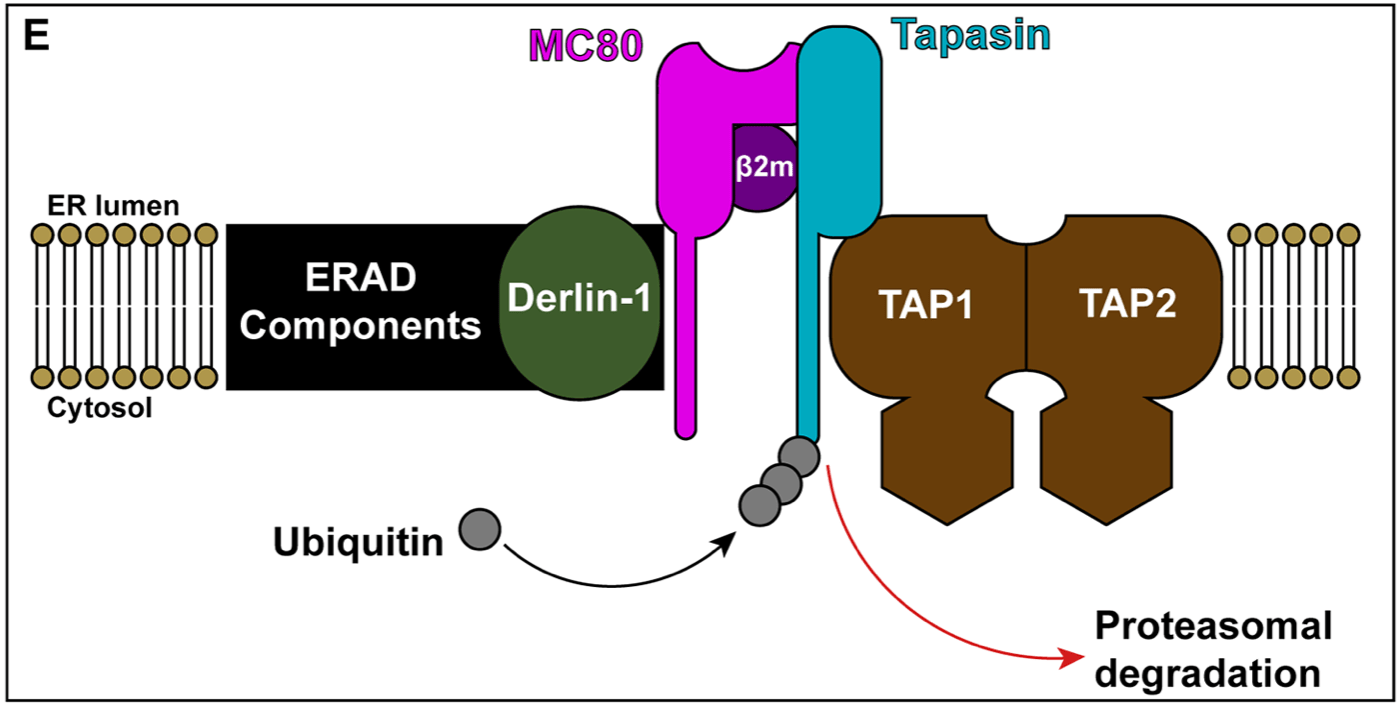
MC80 induces the degradation of Tpn through the ubiquitin-proteasomal pathway
Harvey IB, Wang X, Fremont DH. “Molluscum contagiosum virus MC80 sabotages MHC-I antigen presentation by targeting tapasin for ER-associated degradation.” PLoS Pathog. 2019 Apr 29;15(4):e1007711. doi: 10.1371/journal.ppat.1007711.
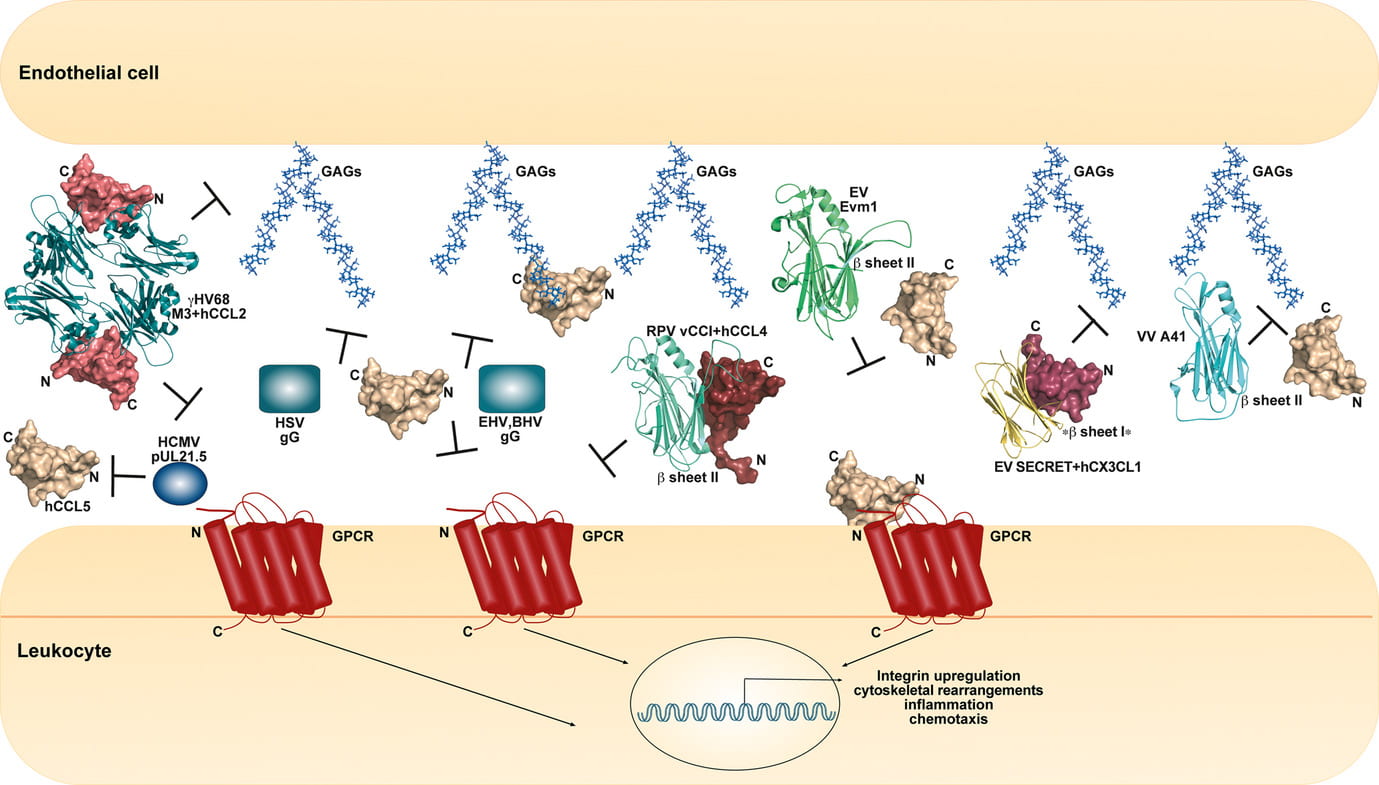
Viral decoy receptor inhibition of chemokines
Epperson ML, Lee CA, Fremont DH. “Subversion of cytokine networks by virally encoded decoy receptors.” Immunol Rev. 2012 Nov;250(1):199-215. doi: 10.1111/imr.12009.
We are also actively studying a number of virally encoded proteins that enable host immune evasion. Large-DNA viruses (i.e. herpesviruses and poxviruses) have evolved a myriad of strategies that include the sabotage of antigen processing, the molecular mimicry of host signaling molecules, and the sequestration of cytokines and chemokines by novel decoy receptors.
Our approach to investigate these systems is based on the production of large quantities of soluble protein in heterologous expression systems, allowing for cognate ligand/ receptor discovery, biophysical interaction analyses, high-resolution structure determinations, and targeted functional studies. Together, these approaches provide a wealth of information on the basic tools with which the immune system operates.